Somatic Variant Interpretation
Introduction
There are a large number of tools and online resources that we use to help interpret cancer variants. We will explore just a few cancer interpretation tools in this section. A more comprehensive list of those that we commonly use is provided below.
Process So Far
Steps below require you to begin with some list of variants in variant call format (.vcf). We will use the final merged, filtered somatic exome VCF from the Somatic SNV and Indel Filtering and Annotation section. Recall that to generate this file, we merged variant calling from the VARSCAN, STRELKA, and MuTect2 programs. All three programs are designed to detect SNVs and small insertions, deletions, and indels. The merged variant file was then left-aligned and trimmed. Then variants were filtered and annotated with VEP.
Additional Filtering
Similar to germline variant interpretation, we can perform additional filtering in the VEP program:
cd /workspace/somatic/
wget https://raw.githubusercontent.com/griffithlab/pmbio.org/master/assets/module_7/V86_38_MUTANTCENSUS-breast.csv
cat V86_38_MUTANTCENSUS-breast.csv | cut -f1 -d , | uniq > breast-ca-gene-list.txt
filter_vep --format vcf -i /workspace/somatic/exome.annotated.vcf -o /workspace/somatic/tumor-exome-clinfilt.vcf --filter "(MAX_AF < 0.01 or not MAX_AF) and FILTER = PASS and SYMBOL in /workspace/somatic/breast-ca-gene-list.txt" --force_overwrite
cat exome.annotated.vcf | grep "^chr" | wc -l
cat tumor-exome-clinfilt.vcf | grep "^chr" | wc -l
-
First, the
cat V86_38_MUTANTCENSUS-breast.csv | cut -f1 -d , | uniq > breast-ca-gene-list.txtreturns inbreast-ca-gene-list.txta unique list of genes found in clinical studies of breast cancer from the Cancer Gene Census (see below). Access to the census through COSMIC is free, but requires a login, so we provide the list for you on the course repository. -
Similar to genomic variant interpretation, the first VEP filter, “MAX_AF <0.01 or not MAX_AF”, returns VCF lines where the variant population frequency is less than 0.01.
-
“FILTER = PASS” selects VCF lines that passed previous quality filters.
-
And SYMBOL in /workspace/somatic/breast-ca-gene-list.txt will limit our results by the list of breast cancer genes from the Cancer Gene Census.
-
Filtering reduced our variants of interest from 1,456 to 51. For a clinical sequencing study, this is a reasonable number of variants to carry forward to manual review.
Highlighted Tools
CRAVAT
CRAVAT (Cancer-Related Analysis of VAriants Toolkit) is a web-based interface for predictive sorting of potenitally pathogenic variants (Paper). To get an overview of our filtered resutls, lets view them in CRAVAT. Access your VCF at http://s#.pmbio.org/somatic/tumor-exome-clinfilt.vcf and then load it into the CRAVAT web interface. Choose “Breast” under the CHASM-3.1 dropdown and submit the report to your email address.
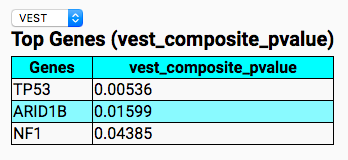
CRAVAT will run two analysis programs CHASM which predicts the functional significance of somatic missense variants, and VEST which is a machine learning algorithm to predict variant pathogenicity.
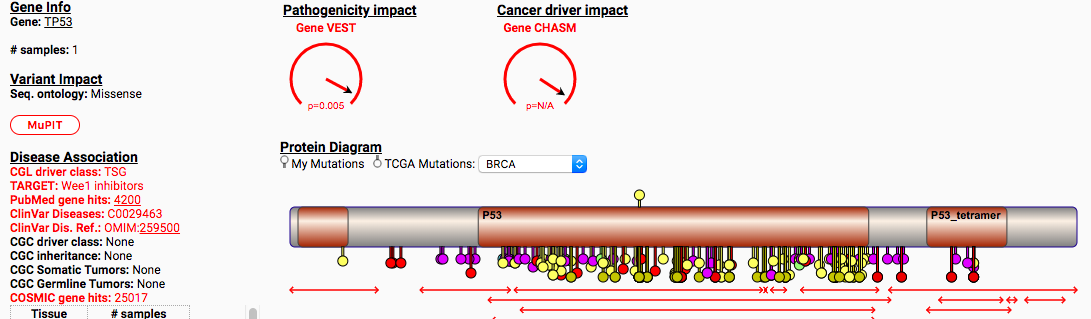
Open the results in the web-based interactive results viewer. Compare the top pathogenic genes by VEST and CHASM. Be sure to check out the additinal tabs for Gene and Variant info as well.
DGIdb
DGIdb (Drug Gene Interaction Database) is drug gene interaction database which can be used to identify inhibitors of activated genes Paper. We could begin investigating our results by passing a list of filtered genes into DGIdb. Like many cancer variant interpretation tools, DGIdb can be quered either through a web interface or at the command line using an application program interface (API). We will try both methods for the VEST-identified pathogenic variants:
First, enter the genes into the web interface as below:
Back on the EC2 instance, call the API:
curl http://dgidb.org/api/v2/interactions.json?genes=TP53,ARID1B,NF1 | python -mjson.tool > dgidb-search.txt
We don’t know exactly what chemotherapy this patient recieved, but typical pre-operative chemotherapy for a triple-negative breast cancer might include doxorubicin and paclitaxel.
cat dgidb-search.txt | grep -E 'DOXORUBICIN|PACLITAXEL'
There is one specific interaction for each drug.
CIViC
CIViC (Clinical Interpretation of Variants in Cancer) is a resource for Clinical Interpreation of Variants in Cancer (WASHU) (Paper).
Exercise
Start with our final list of somatic variants and select a priority set. For example, start with the variants here:
- /workspace/somatic/final/
Additional useful tools and resources for somatic cancer variant interpretation
The following tools are generally applicable to understanding cancer variants. There are hundreds of such tools. These are ones we particularly recommend:
- OncoKB - Annotates oncogenic and predictive/prognositc variants based on expert curation of the literature (Paper)
- VICC knowledgebase aggregator (Variant Interpretation for Cancer Consortium) - A search engine and normalization approach/database that pulls together knowledge from several resources including CIViC and OncoKB.
- CBioPortal - Analysis of large cohorts of cancer data including the Cancer Genome Atlas (TCGA) (Paper)
- ICGC (International Cancer Genome Consortium) - ICGC - A data portal summarizing results from the International Cancer Genome Consortium.
- GDC (Genomic Data Commons Data Portal) - A data portal allowing access to the harmonized analysis results for the TCGA
- [COSMIC](https://cancer.sanger.ac.uk/cosmic (Catalog of Somatic Mutations in Cancer) - Comprehensive database of literature-reported cancer variants
- Cancer Gene Census - Curated list of genes causally implicated in oncogenesis and their assocaited variants (Paper)
- ProteinPaint - A visualization interface for placing an observed amino acid change in the context of protein domains and mutation hotspots according to COSMIC, as well as ClinVar observations.
